Petar Pajic, currently an NSF Postdoctoral Fellow at Yale University (Department of Chemistry), has published a new review article in the journal Genome Biology and Evolution.
Co-authored with Omer Gokcumen (Department of Biological Sciences, University at Buffalo), the paper appeared online on December 24, 2025, in Volume 18, Issue 1 (January 2026), and is available open access under a Creative Commons Attribution License (DOI: 10.1093/gbe/evaf250).
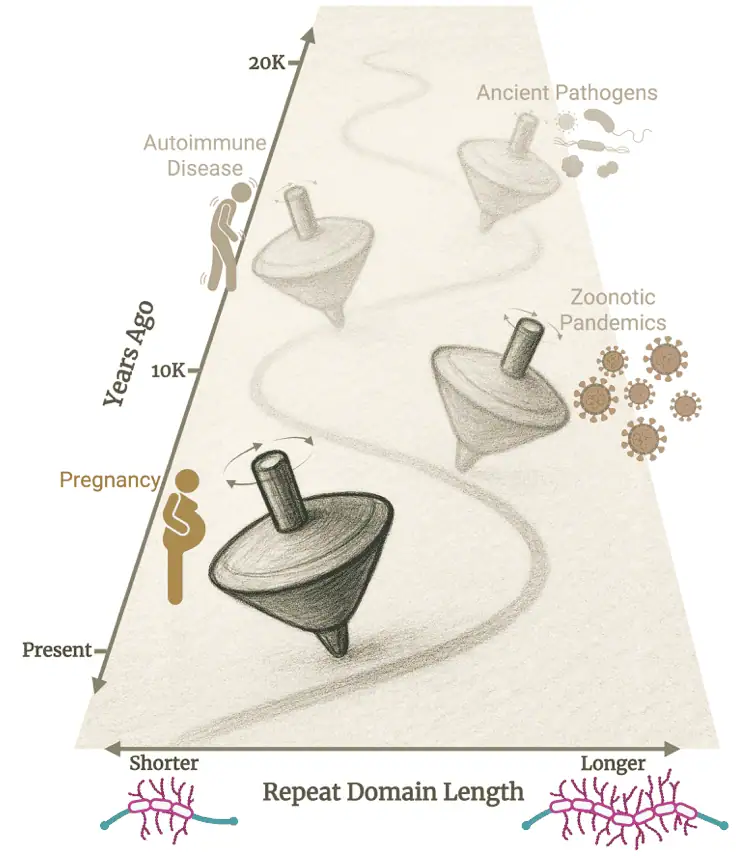
The review explores how variable number tandem repeats (VNTRs) – highly mutable segments of DNA – serve as powerful drivers of evolutionary innovation in mammals, while also carrying significant functional risks. Advances in long-read sequencing technologies have finally allowed accurate mapping and analysis of these previously hard-to-resolve regions, revealing their widespread influence on gene regulation, protein structure variation, and phenotypic diversity.
Key insights include VNTRs’ roles in modulating traits such as skin barrier function (e.g., in the FLG gene), height (e.g., ACAN), behavior (e.g., MAOA), and pathogen defense (e.g., mucin genes). The authors propose a theoretical framework centered on evolutionary tradeoffs: balancing selection and neutral mechanisms maintain VNTR variation, enabling adaptive benefits and genetic novelty, yet the same mutability can lead to disease susceptibility through mechanisms like frameshifts.
This work challenges traditional views that prioritize conserved genomic sequences for function, instead highlighting VNTRs’ contributions to “hidden” genetic variation, missing heritability, adaptation, and complex traits in humans and other mammals.
Congratulations to Petar on this timely synthesis of an emerging and exciting area in evolutionary genomics!